The usage of DisGeNet is as follows:
1. Overview of DisGeNet
2. Search database
3. Fuzzy Search Results
4. Specific research result
5. Download the file
6. FAQ
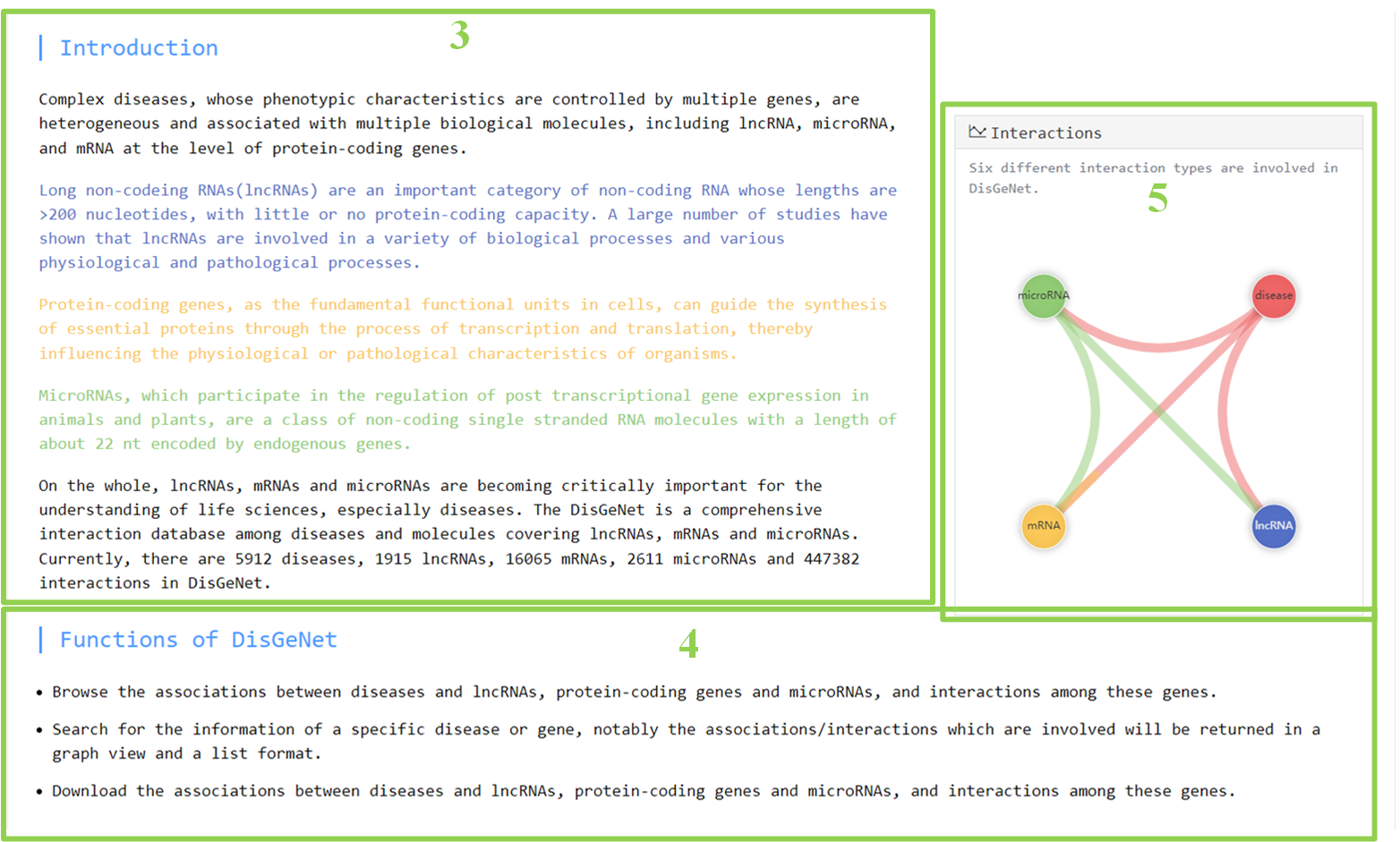
1. Overview of DisGeNet
1.1 Main functions of the database are provided in the menu bar.
1.2 Short search in the menu bar.

1.3 A brief introduction to the database.
1.4 The main functions of the database.
1.5 The main interactions involved in the database.
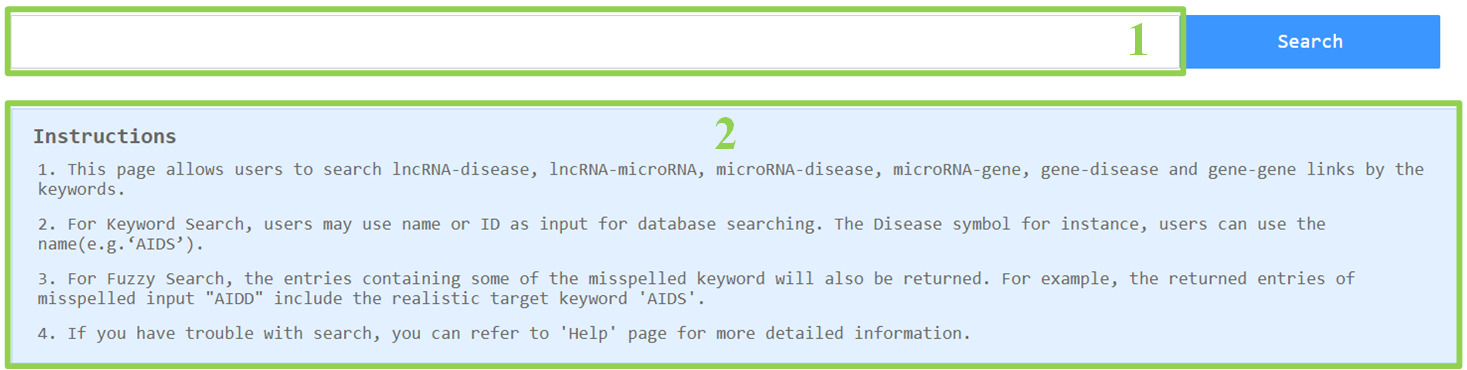
2. Search database
2.1 Input the keyword for the disease, lncRNA, microRNA or mRNA.
2.2 The instructions of the search function.
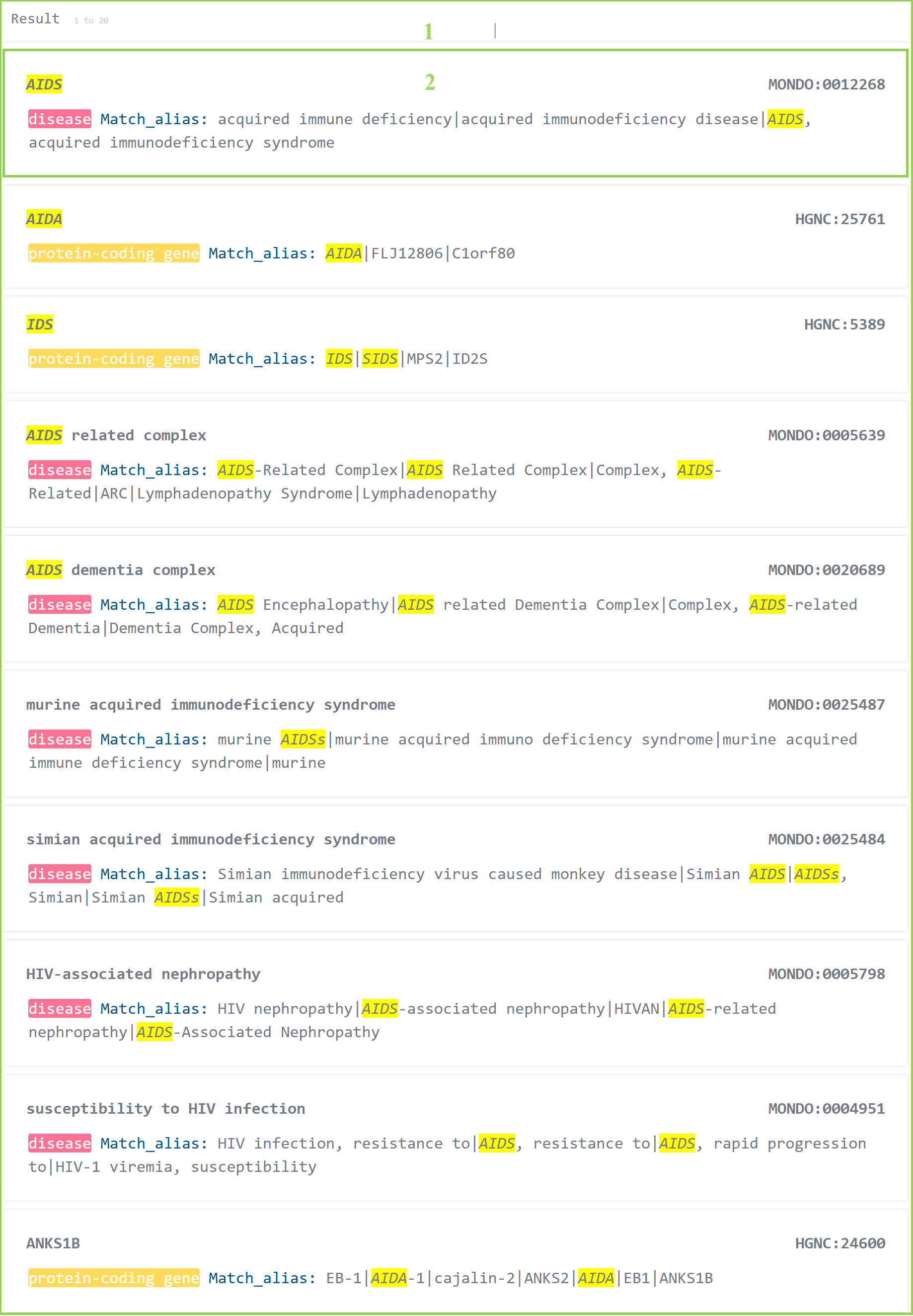
3. Fuzzy Search Results
Fuzzy search is supported in the database, but in order to avoid the abuse of fuzzy search, the top 20 similar results can be displayed in the search results at most.
3.1 The results of fuzzy research.
3.2 Click to select one specific search result.
4. Specific research result
There are two forms, lists and graphs, to represent the results. Besides, all interactions are listed with basic information.
4.1 List Format
4.1.1 Click to link to the View page.
4.1.2 Click to download the search results.
4.1.3 Click to filter interactor type.

4.1.4 Click to display the full informations.
4.1.5 Click to turn the page.
4.1.6 Input the target page number.

4.2 Graph View
Users can move the cursor to a certain position to highlight and get detailed information.
4.2.1 The number of items involved.
4.2.2 The number of interactions involved.
4.2.3 Visual presentation of interactions.
4.2.4 Click to restore or save.

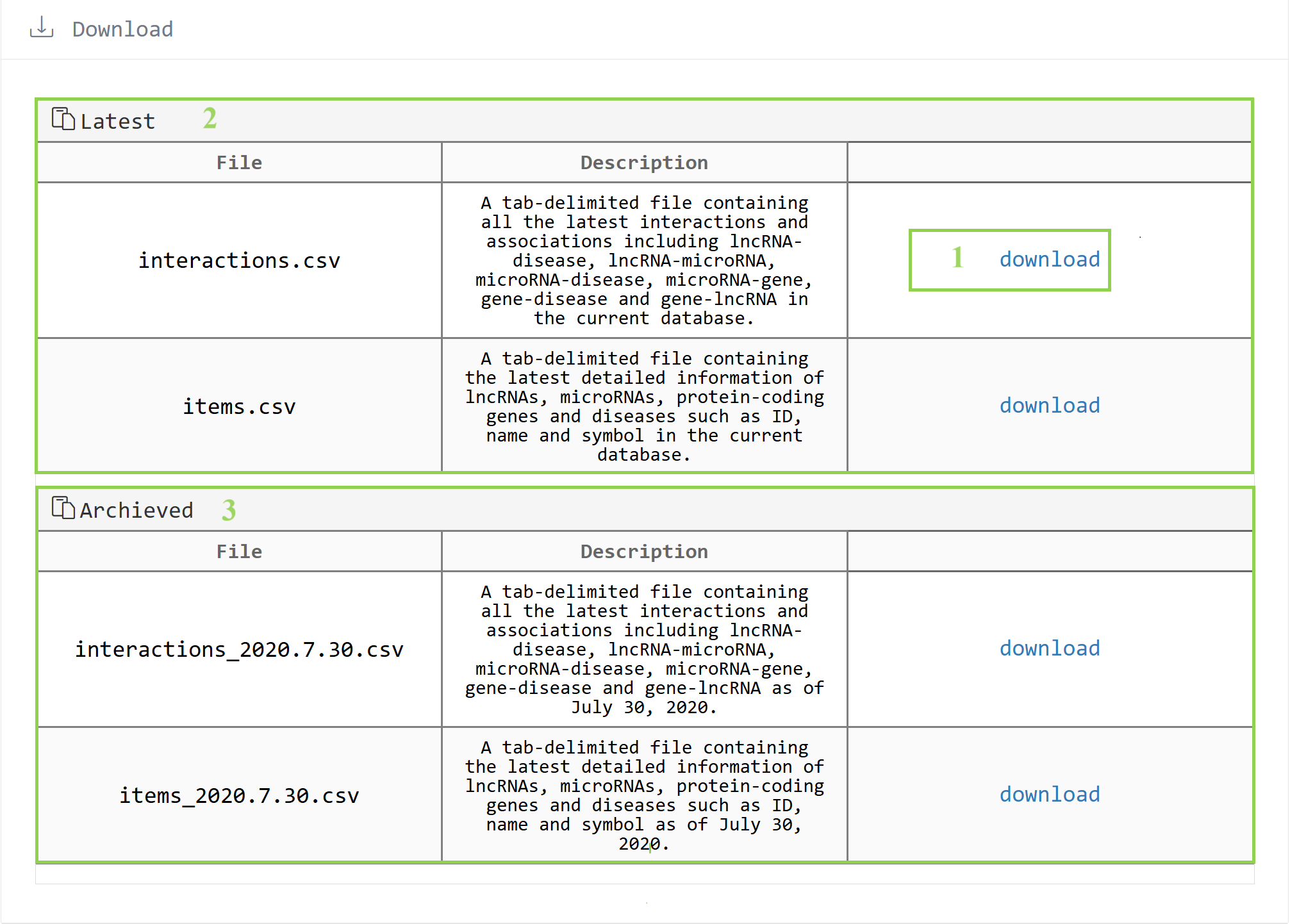
5. Download the file
The files we can download that contain interactions or items are divided into two categories by time point, the latest file and the archived file.
5.1 Click to download the file.
5.2 The latest file.
5.2.3 Instruction of how to use API.
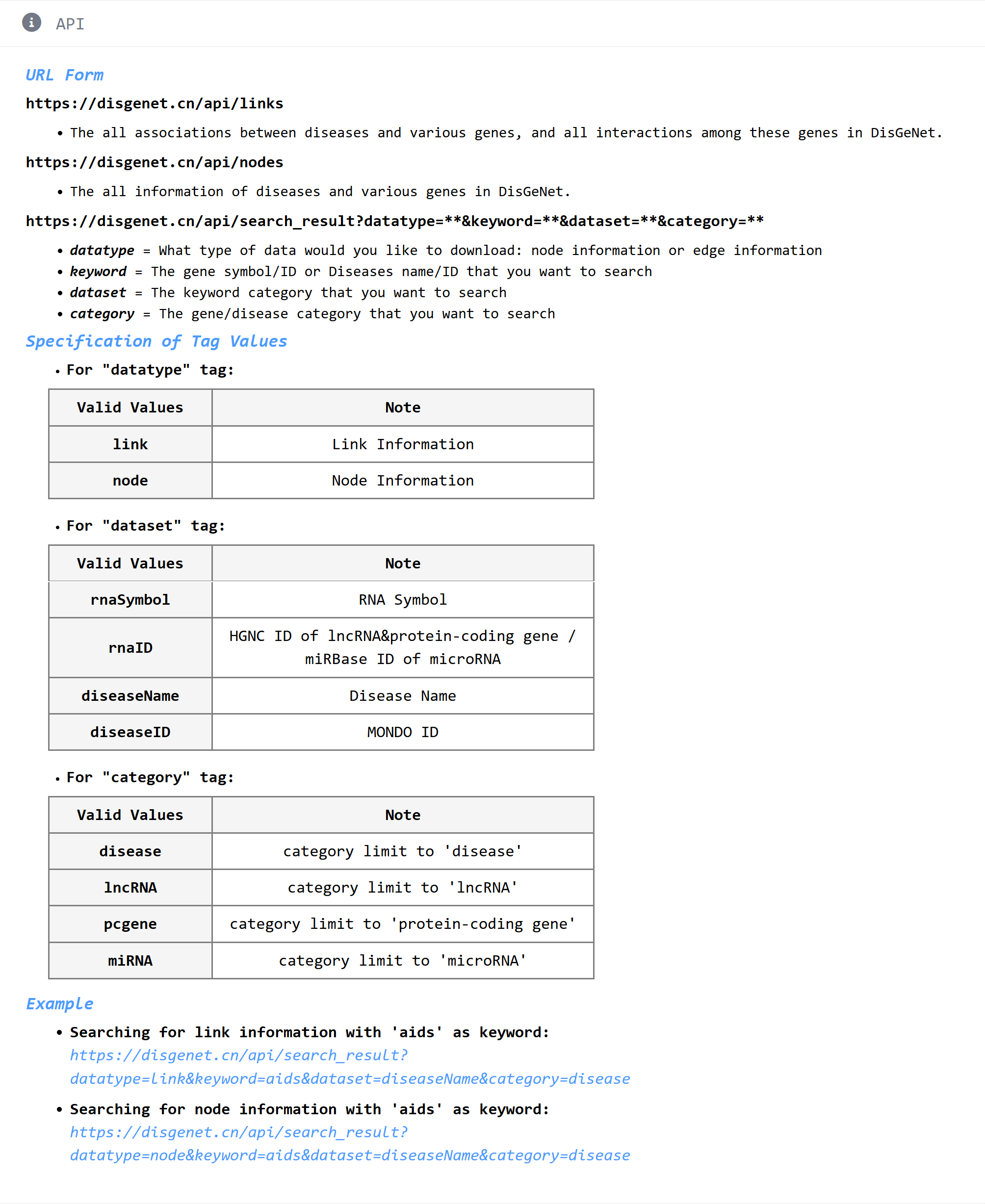
6. FAQ
6.1 How are the RNAs annotated?
For microRNAs, the detailed descriptions are downloaded from miRbase. The general nomenclature of miRNA is "species - microRNA category - serial number". Species are generally represented by three lower case letters, such as hsa, mmu and rno, which represent human, mouse and rat respectively. MicroRNA category refers to pre-miRNA or mature miRNA, which are represented by mirs and miRs respectively. The serial number is an Arabic numeral, representing the discovery sequence of microRNA.
For mRNAs and lncRNAs, annotation file is constructed firstly by integrating information from HGNC, GENCODE and NCBI. Especially to obtain a unified representation, each RNA can be mapped to a term in the file which contained the detailed information.
6.2 Which kind of disease name is used?
The names of diseases come from MONDO, which provides comprehensive information on the integration of DO, Mesh and OMIM.
6.3 What should I do to perform a search?
Just enter the terms for gene symbol or disease name in the search box and click the ‘Search’ button.
The search engine will support the fuzzy search, ignoring the typo in the inputs and providing the similar entries associated with the user’s input. And to prevent the abuse of fuzzy search, only the top 20 most similar items are displayed in the search results.
The searchers are case-insensitive, so a search for “aids”, “aIDS” or “AIDS” will return the same results.
6.4 How to collect and analyze user feedback?
We organize the user feedback interface daily to prevent comment accumulation or disorganization. Additionally, we compile user feedback on a weekly basis, actively engaging with users to gather valuable suggestions. We implement a comprehensive update once a year to ensure that this feedback effectively enhances the utility of the database.